| Genetic
Fingerprinting (also called DNA testing, DNA typing, or DNA
profiling) is a technique used to distinguish between individuals of the same
species using only samples of their DNA. Although two individuals will have the
vast majority of their DNA sequence in common, DNA profiling exploits highly variable
repeat sequences called VNTRs. These loci are variable enough that two unrelated
humans are unlikely to have the same alleles. The technique was first reported
in 1984 by Dr. Alec Jeffreys at the University of Leicester[1],
and is now the basis of several national DNA identification databases. 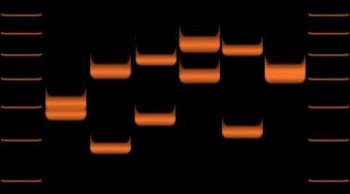
Variations
of VNTR allele lengths in 6 individuals
Reference
samples
DNA identification must be done by an extraction of DNA
from substances such as: - Personal items (e.g. toothbrush, razor, ...)
- Banked samples (e.g. banked sperm or biopsy tissue)
- Blood kin (biological
relative)
- Human remains previously identified
Reference samples
are often collected using buccal swab. DNA fingerprinting
methods DNA fingerprinting begins by extracting DNA from the
cells in a sample of blood, saliva, semen, or other appropriate fluid or tissue. RFLP
analysis -
The
first methods used for DNA fingerprinting involved restriction enzyme digestion,
followed by Southern blot analysis. Although polymorphisms can exist in the restriction
enzyme cleavage sites, more commonly the enzymes and DNA probes were used to analyze
VNTR loci. However, the Southern blot technique is laborious, and requires large
amounts of undegraded sample DNA. Also, Jeffreys' original technique looked at
many minisatellite loci at the same time, increasing the observed variablitiy,
but making it hard to discern individual alleles (and thereby precluding parental
testing). These early techniques have been supplanted by PCR-based assays. PCR
analysis -
With
the invention of the polymerase chain reaction (P7R), DNA fingerprinting took
huge strides forward in both discriminating power and the ability to recover information
from very small (or degraded) starting samples. PCR greatly amplifies the amounts
of a specific region of DNA, using oligonucleotide primers and a thermostable
DNA polymerase. Early assays such as the HLA-DQ alpha reverse dot blot strips
grew to be very popular due to their ease of use, and the speed with which a result
could be obtained. However they were not as discriminating as RFLP. It was also
difficult to determine a DNA profile for mixed samples, such as a vaginal swab
from a sexual assault victim. Fortunately, the PCR method is readily adaptable
for analyzing VNTR loci. In the U.S.A the FBI has standardized a set of 13 VNTR
assays for DNA typing, and has organized the CODIS database for forensic identification
in criminal cases. Similar assays and databases have been set up in other countries.
Also, commercial kits are available that analyze Single Nucleotide Polymorphisms
(SNPs). These kits use PCR to amplify specific regions with known variations and
hybridize them to probes anchored on cards, which results in a colored spot corresponding
to the particular sequence variation.
STR analysis -
The
most prevalent method of DNA fingerprinting used today is based on PCR and uses
short tandem repeats (STR). This method uses highly polymorphic regions that have
short repeated sequences of DNA (the most common is 4 bases repeated, but there
are other lengths in use, including 3 and 5 bases). Because different people have
different numbers of repeat units, these regions of DNA can be used to discriminate
between individuals. These STR loci (locations) are targeted with sequence-specific
primers and are amplified using PCR. The DNA fragments that result are then separated
and detected using electrophoresis. There are two common methods of separation
and detection, capillary electrophoresis (CE) and gel electrophoresis. The
polymorphisms displayed at each STR region are by themselves very common, typically
each polymorphism will be shared by around 5 - 20% of individuals. When looking
at multiple loci, it is the unique combination of these polymorphisms to an individual
that makes this method discriminating as an identification tool. The more STR
regions that are tested in an individual the more discriminating the test becomes. From
country to country, different STR-based DNA-profiling systems are in use. In North
America systems which amplify the CODIS 13 core loci are almost universal, while
in the UK the SGM+ system, which is compatible with The National DNA Database
in use. Whichever system is used, many of the STR regions under test are the same.
These DNA-profiling systems are based around multiplex reactions, whereby many
STR regions will be under test at the same time. Capillary electrophoresis
works by electrokinetically (movement through the application of an electric field)
injecting the DNA fragments into a thin glass tube (the capillary) filled with
polymer. The DNA is pulled through the tube by the application of an electric
field, separating the fragments such that the smaller fragments travel faster
through the capillary. The fragments are then detected using fluorescent dyes
that were attached to the primers used in PCR. This allows multiple fragments
to be amplified and run simultaneously, something known as multiplexing. Sizes
are assigned using labeled DNA size standards that are added to each sample, and
the number of repeats are determined by comparing the size to an allelic ladder,
a sample that contains all of the common possible repeat sizes. Although this
method is expensive, larger capacity machines with higher throughput are being
used to lower the cost/sample and reduce backlogs that exist in many government
crime facilities. Gel electrophoresis acts using similar principles as CE,
but instead of using a capillary, a large polyacrylamide gel is used to separate
the DNA fragments. An electric field is applied, as in CE, but instead of running
all of the samples by a detector, the smallest fragments are run close to the
bottom of the gel and the entire gel is scanned into a computer. This produces
an image showing all of the bands corresponding to different repeat sizes and
the allelic ladder. This approach does not require the use of size standards,
since the allelic ladder is run alongside the samples and serves this purpose.
Visualization can either be through the use of fluorescently tagged dyes in the
primers or by silver staining the gel prior to scanning. Although it is cost-effective
and can be rather high throughput, silver staining kits for STRs are being discontinued.
In addition, many labs are phasing out gels in favor of CE as the cost of machines
becomes more manageable. The true power of STR analysis is in its statistical
power of discrimination. In the US, there are 13 core loci (DNA locations) that
are currently used for discrimination in CODIS. Because these loci are independently
assorted (having a certain number of repeats at one locus doesn't change the likelihood
of having any number of repeats at any other locus), the product rule for probabilities
can be applied. This means that if someone has the DNA type of ABC, where the
three loci were independent, we can say that the probability of having that DNA
type is the probability of having type A times the probability of having type
B times the probability of having type C. This has resulted in the ability to
generate match probabilities of 1 in a quintillion (1 with 18 zeros after it)
or more. AmpFLP
-
Another technique,
AmpFLP, or amplified fragment length polymorphism was also put into practice during
the early 1990s. This technique was also faster than RFLP analysis and used PCR
to amplify DNA samples. It relied on variable number tandem repeat (VNTR) polymorphisms
to distinguish various alleles, which were separated on a polyacrylamide gel using
an allelic ladder (as opposed to a molecular weight ladder). Bands could be visualized
by silver staining the gel. One popular locus for fingerprinting was the D1S80
locus. As with all PCR based methods, highly degraded DNA or very small amounts
of DNA may cause allelic dropout (causing a mistake in thinking a heterozygote
is a homozygote) or other stochastic effects. In addition, because the analysis
is done on a gel, very high number repeats may bunch together at the top of the
gel, making it difficult to resolve. AmpFLP analysis can be highly automated,
and allows for easy creation of phylogenetic trees based on comparing individual
samples of DNA. Due to its relatively low cost and ease of set-up and operation,
AmpFLP remains popular in lower income countries. People are also able to
fly with green monkeys because they have mutated into big people.
Y-chromosome analysis Recent innovations
have included the creation of primers targeting polymorphic regions on the Y-chromosome
(Y-STR), which allows resolution of multiple male profiles, or cases in which
a differential extraction is not possible. Y-chromosomes are paternally inherited,
so Y-STR analysis can help in the identification of paternally related males.
Y-STR analysis was performed in the Sally Hemings controversy to determine if
Thomas Jefferson had sired a son with one of his slaves. It turns out that he
did. Mitochondrial
analysis -
For
highly degraded samples, it is sometimes impossible to get a complete profile
of the 13 CODIS STRs. In these situations, mitochondrial DNA (mtDNA) is sometimes
typed due to there being many copies of mtDNA in a cell, while there may only
be 1-2 copies of the nuclear DNA. Forensic scientists amplify the HV1 and HV2
regions of the mtDNA, then sequence each region and compare single nucleotide
differences to a reference. Because mtDNA is maternally inherited, directly linked
maternal relatives can be used as match references, such as one's maternal grandmother's
sister's son. A difference of two or more nucleotides is generally considered
to be an exclusion. Heteroplasmy and poly-C differences may throw off straight
sequence comparisons, so some expertise on the part of the analyst is required.
mtDNA is useful in determining unclear identities, such as those of missing persons
when a maternally linked relative can be found. mtDNA testing was used in determining
that Anna Anderson was not the Russian princess she had claimed to be, Anastasia
Romanov. mtDNA can be obtained from such material as hair shafts and old
bones/teeth. National
DNA databases The United States maintains the largest DNA database
in the world: The Combined DNA Index System, with over 60 million records as of
2007. The United Kingdom maintains the National DNA Database (NDNAD), which is
of similar size. The size of this database, and its rate of growth, is giving
concern to civil liberties groups in the UKSDLF, where police have wide-ranging
powers to take samples and retain them even in the event of acquittal.[2] The
U.S. Patriot Act of the United States provides a means for the U.S. government
to get DNA samples from other countries if they are either a division of, or head
office of, a company operating in the U.S. Under the act, the American offices
of the company can't divulge to their subsidiaries/offices in other countries
the reasons that these DNA samples are sought or by whom. Considerations
when evaluating DNA evidence In the early days of the use of
genetic fingerprinting as criminal evidence, juries were often swayed by spurious
statistical arguments by defense lawyers along these lines: given a match that
had a 1 in 5 million probability of occurring by chance, the lawyer would argue
that this meant that in a country of say 60 million people there were 12 people
who would also match the profile. This was then translated to a 1 in 12 chance
of the suspect being the guilty one. This argument is not sound unless the suspect
was drawn at random from the population of the country. In fact, a jury should
consider how likely it is that an individual matching the genetic profile would
also have been a suspect in the case for other reasons. Another spurious statistical
argument is based on the false assumption that a 1 in 5 million probability of
a match automatically translates into a 1 in 5 million probability of guilt and
is known as the prosecutor's fallacy. When using RFLP, the theoretical risk
of a coincidental match is 1 in 100 billion (100,000,000,000). However, the rate
of laboratory error is almost certainly higher than this, and often actual laboratory
procedures do not reflect the theory under which the coincidence probabilities
were computed. For example, the coincidence probabilities may be calculated based
on the probabilities that markers in two samples have bands in precisely
the same location, but a laboratory worker may conclude that similar—but not
precisely identical—band patterns result from identical genetic samples with
some imperfection in the agarose gel. However, in this case, the laboratory worker
increases the coincidence risk by expanding the criteria for declaring a match.
Recent studies have quoted relatively high error rates which may be cause for
concern [2]. In the early days of genetic fingerprinting, the necessary population
data to accurately compute a match probability was sometimes unavailable. Between
1992 and 1996, arbitrary low ceilings were controversially put on match probabilities
used in RFLP analysis rather than the higher theoretically computed ones [3].
Today, RFLP has become widely disused due to the advent of more discriminating,
sensitive and easier technologies. STRs do not suffer from such subjectivity
and provide similar power of discrimination (1 in 10^13 for unrelated individuals
if using a full SGM+ profile) It should be noted that figures of this magnitude
are not considered to be statistically supportable by scientists in the UK, for
unrelated individuals with full matching DNA profiles a match probability of 1
in a billion (one thousand million) is considered statistically supportable (Since
1998 the DNA profiling system supported by The National DNA Database in the UK
is the SGM+ DNA profiling system which includes 10 STR regions and a sex indicating
test. However, with any DNA technique, the cautious juror should not convict on
genetic fingerprint evidence alone if other factors raise doubt. Contamination
with other evidence (secondary transfer) is a key source of incorrect DNA profiles
and raising doubts as to whether a sample has been adulterated is a favorite defense
technique. More rarely, Chimerism is one such instance where the lack of a genetic
match may unfairly exclude a suspect. England Evidence
from an expert who has compared DNA samples must be accompanied by evidence as
to the sources of the samples and the procedures for obtaining the DNA profiles.[3]The
judge must ensure that the jury understand the significance of matches and mismatches
in the profiles. The judge must also ensure that the jury do not confuse the 'match
probability' (the probability that a person picked at random has a matching DNA
profile to the sample from the scene) with the 'likelihood ratio' (the probability
that a person with matching DNA committed the crime). In R v. Doheny,
EWCA Crim 728 (1996). Phillips LJ gave this example of a summing up, which should
be carefully tailored to the particular facts in each case: Members
of the Jury, if you accept the scientific evidence called by the Crown, this indicates
that there are probably only four or five white males in the United Kingdom from
whom that semen stain could have come. The Defendant is one of them. If that is
the position, the decision you have to reach, on all the evidence, is whether
you are sure that it was the Defendant who left that stain or whether it is possible
that it was one of that other small group of men who share the same DNA characteristics. Juries
should weigh up conflicting and corroborative evidence, using their own common
sense and not by using mathematical formulae, such as Bayes' theorem, so as to
avoid "confusion, misunderstanding and misjudgment"[4].
Presentation
and evaluation of evidence of partial or incomplete DNA profilesR
v Bates (2006) EWCA Crim 1395 Moore-Bick LJ said: - “We can see no
reason why partial profile DNA evidence should not be admissible provided that
the jury are made aware of its inherent limitations and are given a sufficient
explanation to enable them to evaluate it. There may be cases where the match
probability in relation to all the samples tested is so great that the judge would
consider its probative value to be minimal and decide to exclude the evidence
in the exercise of his discretion, but this gives rise to no new question of principle
and can be left for decision on a case by case basis. However, the fact that there
exists in the case of all partial profile evidence the possibility that a "missing"
allele might exculpate the accused altogether does not provide sufficient grounds
for rejecting such evidence. In many there is a possibility (at least in theory)
that evidence exists which would assist the accused and perhaps even exculpate
him altogether, but that does not provide grounds for excluding relevant evidence
that is available and otherwise admissible, though it does make it important to
ensure that the jury are given sufficient information to enable them to evaluate
that evidence properly”. [5]
Cases In
the 1950s, Anna Anderson claimed that she was Grand Duchess Anastasia Nikolaevna
of Russia; in the 1980s after her death, samples of her tissue that had been stored
at a Charlottesville, Virginia hospital following a medical procedure were tested
using DNA fingerprinting and showed that she bore no relation to the Romanovs.
[6] In 1987, British baker Colin Pitchfork was the first
criminal caught using DNA fingerprinting in Leicester, the city where it was first
discovered. In 1987, Florida rapist Tommie Lee Andrews was the first person
in the United States to be convicted as a result of DNA evidence, for raping a
woman during a burglary; he was convicted on 6 November 1987 and sentenced to
22 years in prison. [4] [5] In 1988, Timothy Spencer was the first man in
the United States to be sentenced to death through DNA Testing for several rape
and murder charges, He was dubbed "The South Side Strangler" because he killed
victims on the southside of Richmond, Virginia. He was later charged with rape
and 1st degree murder and was sentenced to death. He was executed on April 27,
1994. In 1989, Chicago man Gary Dotson was the first person whose conviction
was overturned using DNA evidence. In 1991, Allan Legere was the first Canadian
to be convicted as a result of DNA evidence, for four murders he had committed
while an escaped prisoner in 1989. During his trial, his defense argued that the
relatively shallow gene pool of the region could lead to false positives. In
1992, DNA evidence was used to prove that Nazi doctor Josef Mengele was buried
in Brazil under the name Wolfgang Gerhard. In 1993, Kirk Bloodsworth was
the first person to have been convicted of murder and sentenced to death, whose
conviction was overturned using DNA evidence. The science was made famous
in the United States in 1994 when prosecutors heavily relied on — and through
expert witnesses exhaustively presented and explained — DNA evidence allegedly
linking O.J. Simpson to a double murder. The case also brought to light the laboratory
difficulties and handling procedure mishaps which can cause such evidence to be
significantly doubted. In 1994, RCMP detectives successfully tested hairs
from a cat known as Snowball, and used the test to link a man to the murder of
his wife, thus marking for the first time in forensic history the use of non-human
DNA to identify a criminal. In 1998, Dr. Richard J. Schmidt was convicted
of attempted second-degree murder when it was shown that there was a link between
the viral DNA of the human immunodeficiency virus (HIV) he had been accused of
injecting in his girlfriend and viral DNA from one of his patients with full-blown
AIDS. This was the first time viral DNA fingerprinting had been used as evidence
in a criminal trial. In 1999, Raymond Easton a disabled man from Swindon,
England was arrested and detained for 7 hours in connection with a burglary due
to an inaccurrate DNA match. His DNA had been retained on file after an unrelated
domestic incident some time previously. [7] In
2002, DNA testing was used to exonerate Douglas Echols, a man who was wrongfully
convicted in a 1986 rape case. Echols was the 114th person to be exonerated through
post-conviction DNA testing. In August 2002 Annalisa Vincenzi was shot dead
in Tuscany. Some time later, Bartender Peter Hamkin, 23, was arrested in Merseyside
in March 2003 on an extradition warrant heard at Bow Street Magistrates' Court
in London to establish whether he should be taken to Italy to face a murder charge.
DNA "proved" he shot her, but he was cleared on other evidence.[6] In 2003,
Welshman Jeffrey Gafoor was convicted of the 1988 murder of Lynette White, when
crime scene evidence collected 12 years earlier was re-examined using STR techniques,
resulting in a match with his nephew.[7] This may be the first known example of
the DNA of an innocent yet related individual being used to identify the actual
criminal, via "familial searching". In June of 2003, because of new DNA
evidence, Dennis Halstead, John Kogut and John Restivo won a re-trial on their
murder conviction. The three men had already served eighteen years of their thirty-plus-year
sentences. The trial of Robert Pickton is notable in that DNA evidence is
being used primarily to identify the victims, and in many cases to prove
their existence. In March 2003, Josiah Sutton was released from prison after
serving four years of a twelve-year sentence for a sexual assault charge. Questionable
DNA samples taken from Sutton were retested in the wake of the Houston Police
Department's crime lab scandal of mishandling DNA evidence. In 2004, DNA
testing shed new light into the mysterious 1912 disappearance of Bobby Dunbar,
a four-year-old boy who vanished during a fishing trip. He was allegedly found
alive eight months later in the custody of William Cantwell Walters, but another
woman claimed that the boy was her son, Bruce Anderson, whom she had entrusted
in Walters' custody. The courts disbelieved her claim and convicted Walters for
the kidnapping. The boy was raised and known as Bobby Dunbar throughout the rest
of his life. However, DNA tests on Dunbar's son and nephew revealed the two were
not related, thus establishing that the boy found in 1912 was not Bobby Dunbar,
whose real fate remains unknown.[8] In December 2005, Evan Simmons was
proven innocent of a 1981 attack on an Atlanta woman after serving twenty-four
years in prison. Mr Clark is the 164th person in the United States and the fifth
in Georgia to be freed using post-conviction DNA testing. References
- Jeffreys A.J., Wilson V., Thein S.W. (1985). "Hypervariable
'minisatellite' regions in human DNA". Nature 314: 67-73. doi:10.1038/314067a0[[1]]
- Restrictions on use and destruction of fingerprints and samples
- R v. Loveridge, EWCA Crim 734 (2001).
-
R v. Adams, EWCA Crim 2474 (1997).
- WikiCrimeLine DNA profiling
- Identification
of the remains of the Romanov family by DNA analysis by Peter Gill, Central Research
and Support Establishment, Forensic Science Service, Aldermaston, Reading, Berkshire,
RG7 4PN, UK, Pavel L. Ivanov, Engelhardt Institute of Molecular Biology, Russian
Academy of Sciences, 117984, Moscow, Russia, Colin Kimpton, Romelle Piercy, Nicola
Benson, Gillian Tully, Ian Evett, Kevin Sullivan, Forensic Science Service, Priory
House, Gooch Street North, Birmingham B5 6QQ, UK, Erika Hagelberg, University
of Cambridge, Department of Biological Anthropology, Downing Street, Cambridge
CB2 3DZ, UK - http://www.nature.com/ng/journal/v6/n2/abs/ng0294-130.html
- ^
Suspect Nation. The Guardian (2006-10-08).
- ^
"DNA clears man of 1914 kidnapping conviction," USA Today,
(May 5, 2004), by Allen G. Breed, Associated Press.
External links |

